3' end sequencing protocols
Learn more about the techniques used for 3' end sequencing.
2P-Seq
In the 2P-Seq protocol, reverse transcription is accomplished by an anchored oligo(dT) primer. The products of reverse transcription and PCR amplification are expected to have 20 As preceding the 3' adapter. Libraries are sequenced in anti-sense direction with a custom primer requiring to reverse complement reads to pinpoint the 3' end.

Publications & data sets
Spies, N., Burge, C.B., Bartel, D.P. 3' UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res 23, 2078-90 (2013).

3'-Seq (Mayr)
In the 3'-Seq protocol by Mayr and colleagues, reverse transcription is accomplished by an anchored oligo(dT) primer. The products of reverse transcription and PCR amplification are expected to have 17 As preceding the 3' adapter. Libraries are sequenced in sense direction requiring removal of the 3' adapter sequence and preceding As to pinpoint the 3' end.

Publications & data sets
Lianoglou, S., et al. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev 27, 2380-96 (2013).

Singh, I. et al. Widespread intronic polyadenylation diversifies immune cell transcriptomes. Nat Commun 9, (2018)

Lee, S.-H. et al. Widespread intronic polyadenylation inactivates tumor suppressor genes in leukemia. Nature 561, 127–131 (2018)

- GSM3028273
- GSM3028274
- GSM3028275
- GSM3028276
- GSM3028277
- GSM3028278
- GSM3028279
- GSM3028280
- GSM3028281
- GSM3028282
- GSM3028283
- GSM3028284
- GSM3028285
- GSM3028286
- GSM3028287
- GSM3028288
- GSM3028289
- GSM3028290
- GSM3028291
- GSM3028292
- GSM3028293
- GSM3028294
- GSM3028295
- GSM3028296
- GSM3028297
- GSM3028298
- GSM3028299
- GSM3028300
- GSM3028301
- GSM3039795
- GSM3039796
- GSM3039797
- GSM3039798
- GSM3039799
- GSM3039800
- GSM3039801
- GSM3039802
- GSM3039803
- GSM3039804
- GSM3039805
- GSM3039806
- GSM3039807
- SRX351949
- SRX351950
- SRX351952
- SRX351953
- SRX359328
- SRX359329
- SRX359330
- SRX359331
- SRX359332
- SRX359333
- SRX359334
- SRX359335
- SRX359336
- SRX359337
- SRX359339
- SRX359340
- SRX359341
3'READS
3' region extraction and deep sequencing (3'READS) is a protocol that utilizes a special oligo(dT) primer ( 45 thymidines followed by 5 uridines) to caputre poly(A) containing RNA fragments. After partial digestion of the poly(A) tail, the fragments are subjected to adapter ligation, reverse transcription, and PCR amplification before they are sequenced in anti-sense direction. The cleavage site is inferred as the first non-A of the 3' end of the read's reverse complement.

Publications & data sets
Li, W. et al. Systematic Profiling of poly(A)+ Transcripts Modulated by Core 3’ End Processing and Splicing Factors Reveals Regulatory Rules of Alternative Cleavage and Polyadenylation. PLoS Genetics 11 (4), e1005166 (2015).

Yang, Y. et al. PAF Complex Plays Novel Subunit-Specific Roles in Alternative Cleavage and Polyadenylation. PLoS Genet 12, (2016)

Li, W. et al. Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. BMC Biol 14, (2016)

Zheng, D. et al. Cellular stress alters 3′UTR landscape through alternative polyadenylation and isoform-specific degradation. Nat Commun 9, (2018)

Wang, R. et al. A compendium of conserved cleavage and polyadenylation events in mammalian genes. Genome Res 10, 1427-1441. (2018)

Publicly available data without publication.

- GSM1518071
- GSM1518072
- GSM1518073
- GSM1518074
- GSM1518075
- GSM1518076
- GSM1518077
- GSM1518078
- GSM1518079
- GSM1518080
- GSM1518081
- GSM1518082
- GSM1518083
- GSM1518084
- GSM1518085
- GSM1518086
- GSM1518087
- GSM1518088
- GSM1518089
- GSM1518090
- GSM1518091
- GSM1518092
- GSM1518093
- GSM1518094
- GSM1518095
- GSM1518096
- GSM1518097
- GSM1518098
- GSM1518099
- GSM1518100
- GSM1518101
- GSM1518102
- GSM1518103
- GSM1518104
- GSM1518105
- GSM1518106
- GSM1518107
- GSM1518108
- GSM1518109
- GSM1518110
- GSM1518111
- GSM1518112
- GSM1518113
- GSM1518114
- GSM1586363
- GSM1586364
- GSM1586365
- GSM1586366
- GSM1586367
- GSM1586368
- GSM1846071
- GSM1846072
- GSM1846073
- GSM1865359
- GSM1865360
- GSM1865361
- GSM1865362
- GSM1865363
- GSM1865364
- GSM1865365
- GSM1906926
- GSM1906927
- GSM1906928
- GSM1906929
- GSM1906930
- GSM1906931
- GSM2717200
- GSM2717201
- GSM2717202
- GSM2717203
- GSM2717204
- GSM2717205
- GSM2717206
- GSM2717207
- GSM2717208
- GSM2717209
- GSM2717222
- GSM2717223
- GSM2717224
- GSM2717225
- GSM2717226
- GSM2717227
- GSM2717228
- GSM2717229
- GSM3022814
- GSM3022815
- GSM3022816
- GSM3022817
- GSM3022818
- GSM3022819
- GSM3022820
- GSM3022821
- GSM3022822
- GSM3022823
- GSM3022824
- GSM3022825
- GSM3022826
- GSM3022827
- GSM3022828
- GSM3375361
- GSM3375362
- GSM3375363
- GSM3375364
- GSM3375365
- GSM3375366
- GSM3375367
- GSM3375368
- GSM3375369
- GSM3375370
- GSM3375371
- GSM3375372
- GSM3375373
- GSM3375374
- GSM3375375
- GSM3375376
- GSM3375377
- GSM3375378
- GSM3375379
- GSM3375380
- GSM3375381
- GSM3375382
- GSM3375386
- GSM3375390
- GSM3375391
- GSM3375392
- GSM3375393
- GSM3375394
- GSM3375395
- GSM3375396
- GSM3375404
- GSM3375405
- GSM3375406
- GSM3375407
- GSM3375408
- GSM3375413
- GSM3375414
- GSM3375419
- GSM3375420
- GSM3375421
- GSM3375423
- GSM3375424
- GSM3375425
- GSM3375426
- GSM3375427
- GSM3375428
- GSM3375429
- GSM3375430
- GSM3375431
3P-Seq
In the 3P-Seq protocol, a biotinylated adapter is ligated to the end of the poly(A) tail via splint-ligation and the poly(A) region is reverse transcribed with only dTTP. After removal of most of the poly(A) tail, adapters are ligated prior to reverse transcription and PCR amplification. Finally, the libraries are sequenced in anti-sense direction and the resulting reads are reverse complemented in order to pinpoint the 3' ends.

Publications & data sets
Jan, C. H., Friedman, R. C., Ruby, J. G. & Bartel, D. P. Formation, regulation and evolution of Caenorhabditis elegans 3’UTRs. Nature 469, 97–101 (2011).

Nam, J. W. et al. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol Cell 53, 1031-43 (2014).

A-seq
In the A-seq protocol, reverse transcription is accomplished by an anchored oligo(dT) primer. The products of reverse transcription and PCR amplification are expected to have six As preceding the 3' adapter. Libraries are sequenced in sense direction requiring removal of the 3' adapter sequence and preceding As to pinpoint the 3' end.

Publications & data sets
Martin, G., Gruber, A. R., Keller, W. & Zavolan, M. Genome-wide analysis of pre-mRNA 3’ end processing reveals a decisive role of human cleavage factor I in the regulation of 3' UTR length. Cell Rep 1, 753–763 (2012).

Gruber, A. R., Martin, G., Keller, W. & Zavolan, M. Cleavage factor Im is a key regulator of 3’ UTR length. RNA Biol 9, 1405–1412 (2012).

Gruber, AR. et al. Global 3’ UTR Shortening Has a Limited Effect on Protein Abundance in Proliferating T Cells. Nat Commun 5, 5465 (2014).

DRS
In the direct RNA sequencing (DRS) protocol, 3' ends of transcripts are hybridized to poly(dT)-coated flow cell surfaces where antisense strand synthesis is initiated. This has the advantage that no prior reverse transcription or cDNA amplification is needed.

Publications & data sets
Yao, C. et al. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc Natl Acad Sci U S A 109, 18773–18778 (2012).

Ji, X. et al. αCP Poly(C) Binding Proteins Act as Global Regulators of Alternative Polyadenylation. Mol Cell Biol 33, 2560–2573 (2013).

Lackford, B. et al. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J (2014).

Rehfeld, A. et al. Alternative polyadenylation of tumor suppressor genes in small intestinal neuroendocrine tumors. Front Endocrinol (Lausanne) 5, 46 (2014).

PAPERCLIP
To capture RNA 3'ends PAPERCLIP takes advantage of the specific interaction of poly(A)-binding protein (PABP) with the RNA's poly(A) tail. After crosslinking RNA-binding proteins to RNAs, the RNAs are fragmented, and RNA fragments crosslinked to PABP are purified with a specific antibody. The protein is then degraded and the RNA fragments are subjected to reverse transcription with a primer that includes sequencing adaptors. Subsequently the cDNA is circularized, the insert is amplified by PCR using primers specific for the adaptor sequences, and finally sequenced in sense direction.
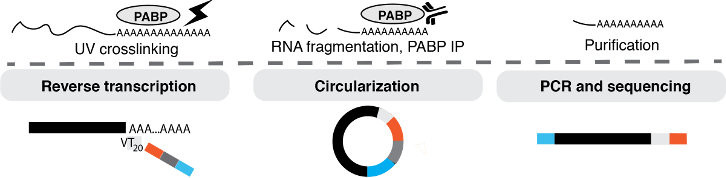
Publications & data sets
Hwang, H.W. et al. PAPERCLIP Identifies MicroRNA Targets and a Role of CstF64/64tau in Promoting Non-canonical poly(A) Site Usage. Cell Rep 15, 423-35 (2016)

Hwang, H.-W. et al. cTag-PAPERCLIP Reveals Alternative Polyadenylation Promotes Cell-Type Specific Protein Diversity and Shifts Araf Isoforms with Microglia Activation. Neuron 95, 1334-1349.e5 (2017)

Jereb, S. et al. Differential 3’ Processing of Specific Transcripts Expands Regulatory and Protein Diversity Across Neuronal Cell Types. Elife 7, (2018)

- GSM1614163
- GSM1614164
- GSM1614165
- GSM1614166
- GSM1614171
- GSM1614172
- GSM1614173
- GSM1614174
- GSM1614175
- GSM1614176
- GSM1857615
- GSM1857616
- GSM1857617
- GSM1857618
- GSM1857619
- GSM1857620
- GSM1857621
- GSM1857622
- GSM1857623
- GSM1857624
- GSM1857625
- GSM1857626
- GSM2467568
- GSM2467569
- GSM2467570
- GSM2467571
- GSM2467572
- GSM2467573
- GSM2467574
- GSM2467575
- GSM2467576
- GSM2467577
- GSM2467578
- GSM2467579
- GSM2467580
- GSM2467581
- GSM2467582
- GSM2901339
- GSM2901340
- GSM2901341
- GSM2901342
- GSM2901343
- GSM2901344
- GSM2901345
- GSM2901346
- GSM2901347
- GSM2901348
- GSM2901349
- GSM2901350
- GSM2901351
PAS-Seq
In the PAS-Seq protocol, reverse transcription is accomplished by an anchored oligo(dT) primer. The products of reverse transcription and PCR amplification are expected to have 20 As preceding the 3' adapter. Libraries are sequenced in anti-sense direction with a costum primer requiring to reverse complement reads to pinpoint the 3' end.

Publications & data sets
Shepard, P. J. et al. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17, 761–772 (2011).

PAT-seq
For PAT-seq, adenylated RNA is extended by dNTPs using Klenow polymerase and a biotinylated anchor-oligo. Internal priming is avoided by a requirement for this 3' extension in subsequent steps. Tagged RNA is fragmented with RNase T1, the 3' end fragments are collected with streptavidin beads and ligated to a 5' linker. After reverse transcription fragments are selected by size to ensure that sequencing reaches the poly(A) tail. Subsequently they are PCR amplified and sequenced in sense direction, requiring removal of 3' As to pinpoint the 3' ends.

Publications & data sets
Elewa, A. et al. POS-1 Promotes Endo-mesoderm Development by Inhibiting the Cytoplasmic Polyadenylation of neg-1 mRNA. Dev Cell 34, 108–118 (2015).

PolyA-seq
Library preparation for the PolyA-seq protocol requires (1) Reverse transcription, primed with an oligo(dT) sequence succeeding an universal sequence used as PCR anchor, (2) second strand synthesis with random hexamers linked to a second PCR primer, and (3) PCR amplification. Sequencing is accomplished in anti-sense orientation with a primer ending in 10 Ts; the resulting reads need to be reverse complemented in order to pinpoint the mRNA cleavage site.

Publications & data sets
Derti, A. et al. A quantitative atlas of polyadenylation in five mammals. Genome Res 22, 1173–1183 (2012).

Batra, R. et al. Loss of MBNL Leads to Disruption of Developmentally Regulated Alternative Polyadenylation in RNA-Mediated Disease. Mol Cell 56, 311–322 (2014)

QuantSeq_REV
QuantSeq is the method implemented by Lexogen, starting from total RNA and requiring a turnaround time of a few hours. The reverse transcription of polyadenylated RNAs is done with an oligo(dT) primer that contains the 3’ adaptor. Second strand synthesis is then carried out by random priming, with oligos that also contain the 5’ adaptor. The library is amplified and sequenced, in the case of QuantSeq REV starting from the last nucleotide of the mRNA in antisense direction.

Publications & data sets
Publicly available data without publication.

- SAMEA4444111
- SAMEA4444112
- SAMEA4444113
- SAMEA4444114
- SAMEA4444115
- SAMEA4444116
- SAMEA4444117
- SAMEA4444118
- SAMEA4444119
- SAMEA4444120
- SAMEA4444121
- SAMEA4444122
- SAMEA4444123
- SAMEA4444124
- SAMEA4444125
- SAMEA4444126
- SAMEA4444127
- SAMEA4444128
- SAMEA4444129
- SAMEA4444130
- SAMEA4444131
- SAMEA4444132
- SAMEA4444133
- SAMEA4444134
- SAMEA4444135
- SAMEA4444136
- SAMEA4444137
SAPAS
In the SAPAS protocol, reverse transcription is accomplished by an anchored oligo(dT) primer. The products of reverse transcription and PCR amplification are expected to have the sequence AAAAAAGAAAAAAGAAAAAA preceding the 3' adapter. Libraries are sequenced in anti-sense direction with a regular primer requiring to trimm 20 nucleotides from the 5' end of reads and to reverse complement reads to pinpoint the 3' end.

Publications & data sets
Fu, Y. et al. Differential genome-wide profiling of tandem 3’ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res 21, 741–747 (2011).

You, L. et al. APASdb: A Database Describing Alternative poly(A) Sites and Selection of Heterogeneous Cleavage Sites Downstream of poly(A) Signals. Nucl Acids Res 43 (Database issue), D59–67 (2015).

- SRX026582
- SRX026583
- SRX026584
- SRX480169
- SRX480179
- SRX480205
- SRX480212
- SRX480221
- SRX480227
- SRX480229
- SRX480250
- SRX480287
- SRX517313
- SRX517314
- SRX517315
- SRX517316
- SRX517317
- SRX517318
- SRX517319
- SRX517320
- SRX517321
- SRX517322
- SRX517323
- SRX517324
- SRX517325
- SRX517326
- SRX517327
- SRX517328
- SRX517329
- SRX517330
- SRX517331
- SRX517332
- SRX517333
- SRX517334